Ancestry#
Output Directory: results/variantAnalysis/ancestry/
Rules
variantAnalysis.smk
AncestryInformativeMarkers
Introduction
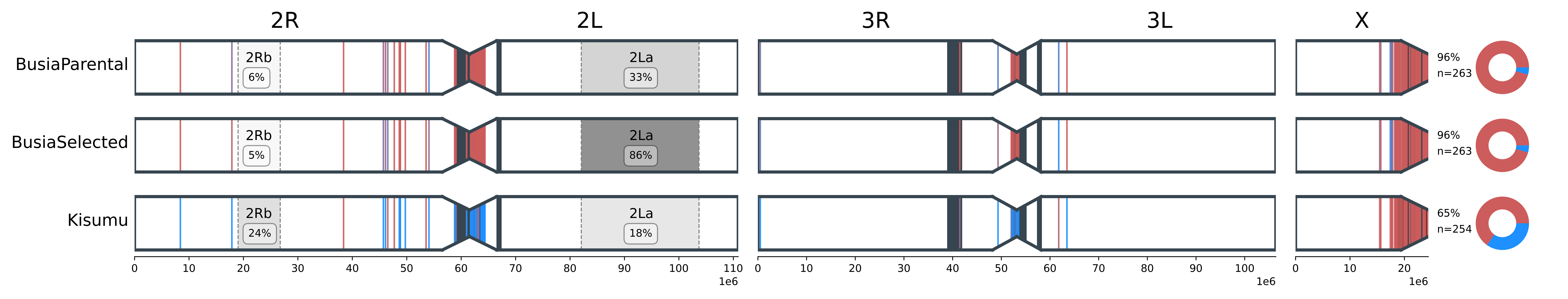
The Anopheles gambiae species complex is a group of closely related sibling species. These species diverged relatively recently, and so they are still capable of hybridising and producing viable offspring. This process is highly relevant from a vector control perspective, as they allow beneficial alleles, such as those under selection due to insecticide resistance, to pass between species. For example, both the L995F kdr mutation and the G280S ace-1 mutation have passed between An. gambiae and An. coluzzii.
One approach to studying ancestry of individuals in the Anopheles gambiae complex is to use ancestry informative markers (AIMs). AIMs are SNPs which show fixed differences (or almost fixed) between species, and thus can be used as diagnostic markers to determine an individuals ancestry. By genotyping AIMs across the genome, we can identify regions of the genome that have introgressed from another species, or identify contamination events that have occured in the insectaries or during sample and library preparation.
RNA-Seq-Pop can use two sets of AIMs, gambiae vs coluzzii, or An. gambiae/coluzzii vs arabiensis.
Results
# the image wouldnt open in markdown
from IPython.display import display, Image
display(Image(filename='../../figures/chromosome_aim_karyo2.png'))